Novel deep learning models in medical imaging appear one after another. The thing that these models still significantly lack is the ability to generalize to unseen clinical data. Unseen data refer to real-life conditions that are typically different from the ones encountered during training. So when we want to apply a model in clinical practice, we are likely to fail. Furthermore, the provided training data is often limited. This constricts the expressive capability of deep models, as their performance is bounded by the number of data. And the only solution is to find more data. Since it is not always possible to find the exact supervised data you want, you may consider transfer learning as a choice.
Transfer learning will be the next driver of ML success ~ Andrew Ng, NeurIPS 2016 tutorial
In natural images, we always use the available pretrained models. We may use them for image classification, object detection, or segmentation. And surprisingly it always works quite well. This mainly happens because RGB images follow a distribution. The shift between different RGB datasets is not significantly large. Admittedly, medical images are by far different. Each medical device produces images based on different physics principles. Subsequently, the distribution of the different modalities is quite dissimilar. The reason we care about it?
Transfer learning of course! If you want to learn the particularities of transfer learning in medical imaging, you are in the right place.
To dive deeper into how AI is used in Medicine, you can’t go wrong with the AI for Medicine online course, offered by Coursera. If you want to focus on medical image analysis with deep learning, I highly recommend starting from the Pytorch-based Udemy Course.
What is Transfer Learning?
Let’s say that we intend to train a model for some task X (domain A). Thus, we assume that we have acquired annotated data from domain A. A task is our objective, image classification, and the domain is where our data is coming from. In medical imaging, think of it as different modalities. When we directly train a model on domain A for task X, we expect it to perform well on unseen data from domain A. What happens if we want to train a model to perform a new task Y?
In transfer learning, we try to store this knowledge gained in solving a task from the source domain A and apply it to another domain B.
The source and target task may or may not be the same. In general, we denote the target task as Y. We store the information in the weights of the model. Instead of random weights, we initialize with the learned weights from task A. If the new task Y is different from the trained task X then the last layer (or even larger parts of the networks) is discarded.
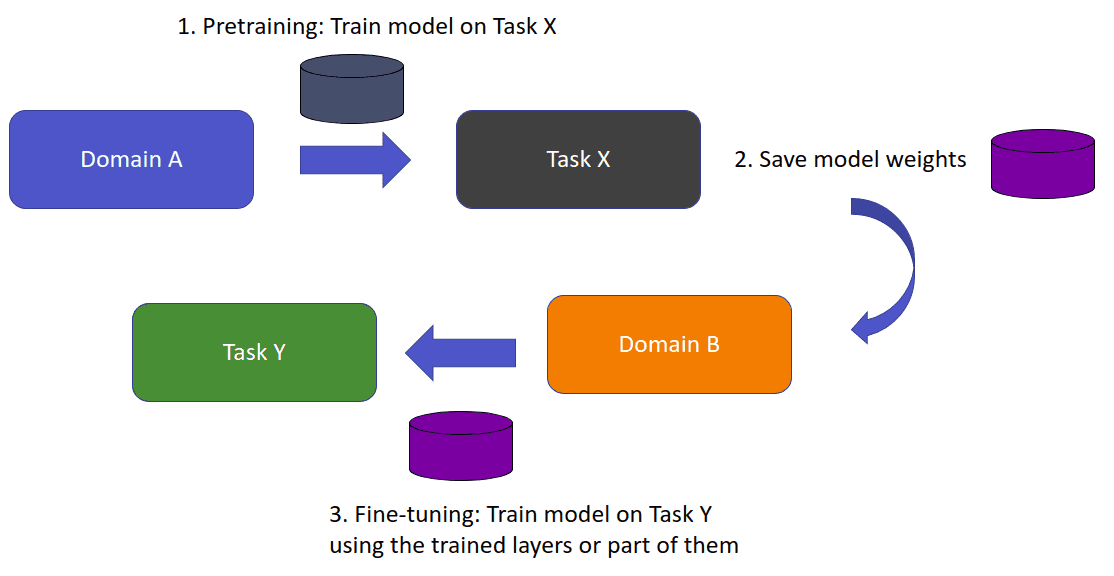 An overview of transfer learning. Image by Author
An overview of transfer learning. Image by Author
For example, for image classification we discard the last hidden layers. In encoder-decoder architectures we often pretrain the encoder in a downstream task. But how different can a domain be in medical imaging? What kind of tasks are suited for pretraining? What parts of the model should be kept for fine tuning?
We will try to tackle these questions in medical imaging.
Keep in mind, that for a more comprehensive overview on AI for Medicine we highly recommend our readers to try this course.
Transfer learning from ImageNet for 2D medical image classification (CT and Retina images)
Obviously, there are significantly more datasets of natural images. The most common one for transfer learning is ImageNet, with more than 1 million images. Therefore, an open question arises: How much ImageNet feature reuse is helpful for medical images?
Let’s introduce some context. ImageNet has 1000 classes. That’s why pretrained models have a lot of parameters in the last layers on this dataset. On the other hand, medical image datasets have a small set of classes, frequently less than 20. So, the design is suboptimal and probably these models are overparametrized for the medical imaging datasets.
In the case of the work that we‘ll describe we have chest CT slices of 224x224 (resized) that are used to diagnose 5 different thoracic pathologies: atelectasis, cardiomegaly, consolidation, edema, and pleural effusion. The RETINA dataset consists of retinal fundus photographs, which are images of the back of the eye.
 A normal fundus photograph of the right eye. Taken from Wikipedia
A normal fundus photograph of the right eye. Taken from Wikipedia
Such images are too large (i.e. 3 x 587 × 587) for a deep neural network.
It is obvious that this 3-channel image is not even close to an RGB image. To understand the impact of transfer learning, Raghu et al [1] introduced some remarkable guidelines in their work: “Transfusion: Understanding Transfer Learning for Medical Imaging”.
In general, one of the main findings of [1] is that transfer learning primarily helps the larger models, compared to smaller ones. Intuitively, it makes sense! Smaller models do not exhibit such performance gains. Moreover, for large models, such as ResNet and InceptionNet, pretrained weights learn different representations than training from random initialization. Apart from that, large models change less during fine-tuning, especially in the lowest layers.
To address these issues, the Raghu et al [1] proposed two solutions:
- Transfer the scale (range) of the weights instead of the weights themselves. This offers feature-independent benefits that facilitate convergence. In particular, they initialized the weights from a normal distribution . The mean and the variance of the weight matrix is calculated from the pretrained weights. This calculation was performed for each layer separately. As a result, the new initialization scheme inherits the scaling of the pretrained weights but forgets the representations. The following plots illustrate the pre-described method (Mean Var) and it’s speedup in convergence.
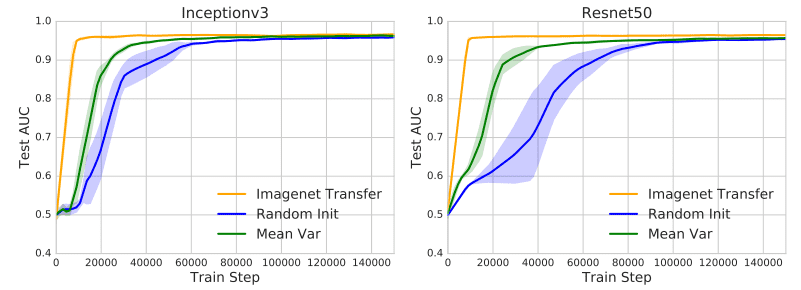 The effect of ImageNet pretraining. Image by [1][source](https://arxiv.org/abs/1902.07208)
The effect of ImageNet pretraining. Image by [1][source](https://arxiv.org/abs/1902.07208)
- Use the pretrained weights only from the lowest two layers. The rest of the network is randomly initialized and fine-tuned for the medical imaging task. This hybrid method has the biggest impact on convergence. To summarize, most of the most meaningful feature representations are learned in the lowest two layers.
Finally, keep in mind that so far we refer to 2D medical imaging tasks. What about 3D medical imaging datasets?
Transfer Learning for 3D MRI Brain Tumor Segmentation
Wacker et al. [4] attempt to use ImageNet weight with an architecture that combines ResNet (ResNet 34) with a decoder. In this way, they simply treat three MRI modalities as RGB input channels of the
pretrained encoder architecture. The pretrained convolutional layers of ResNet used in the downsampling path of the encoder, forming a U-shaped architecture for MRI segmentation.
To process 3D volumes, they extend the 3x3 convolutions inside ResNet34 with 1x3x3 convolutions. Thereby, the number of parameters is kept intact, while pretrained 2D weights are loaded. Simply, the ResNet encoder simply processes the volumetric data slice-wise. They used the Brats dataset where you try to segment the different types of tumors. The different tumor classes are illustrated in the Figure below.
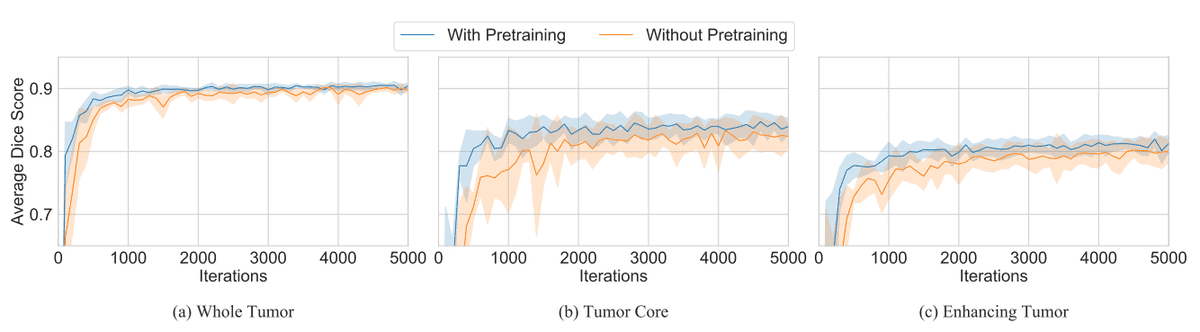 Image by Wacker et al. [4]. Source
Image by Wacker et al. [4]. Source
The results of the pretraining were rather marginal. Moreover, this setup can only be applied when you deal with exactly three modalities. However, this is not always the case.
Transfer Learning for 3D lung segmentation and pulmonary nodule classification
In general, 10%-20% of patients with lung cancer are diagnosed via a pulmonary nodule detection. According to Wikipedia [6]: “A lung nodule or pulmonary nodule is a relatively small focal density in the lung. It is a mass in the lung smaller than 3 centimeters in diameter. The nodule most commonly represents a benign tumor, but in around 20% of cases, it represents malignant cancer.”
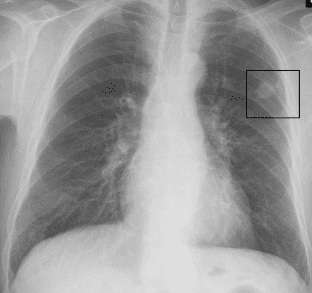 Pulmonary nodule detection. The image is taken from [Wikipedia](Med3D: Transfer Learning for 3D Medical Image Analysis).
Pulmonary nodule detection. The image is taken from [Wikipedia](Med3D: Transfer Learning for 3D Medical Image Analysis).
Let’s go back to our favorite topic. So, if transferring weights from ImageNet is not that effective why don’t we try to add up all the medical data that we can find? Simple, but effective! Chen et al. collected a series of public CT and MRI datasets. Most of the data can be found on Medical Image Decathlon. Nonetheless, the data come from different domains, modalities, target organs, pathologies. They use a family of 3D-ResNet models in the encoder part. The decoder consists of transpose convolutions to upsample the feature in the dimension of the segmentation map. To deal with multiple datasets, different decoders were used. The different decoders for each task are commonly referred to as “heads” in the literature. The depicted architecture is called Med3D. Below you can inspect how they transfer the weights for image classification.
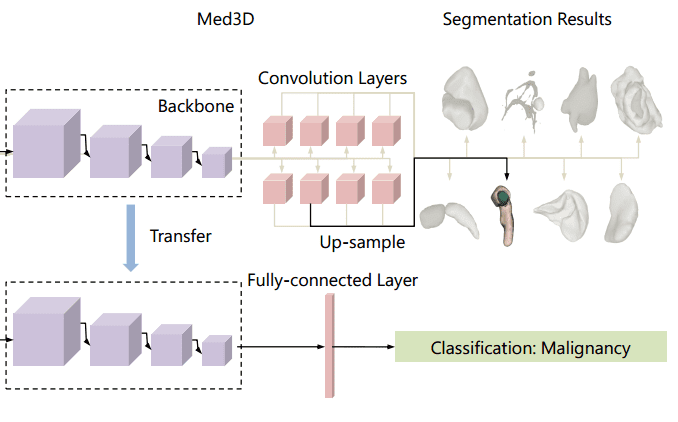 An overview of the Med3D architecture [2]
An overview of the Med3D architecture [2]
To deal with multi-modal datasets they used only one modality. They compared the pretraining on medical imaging with Train From Scratch (TFS) as well as from the weights of the Kinetics, which is an action recognition video dataset. I have to say here, that I am surprised that such a dataset worked better than TFS! The results are much more promising, compared to what we saw before.
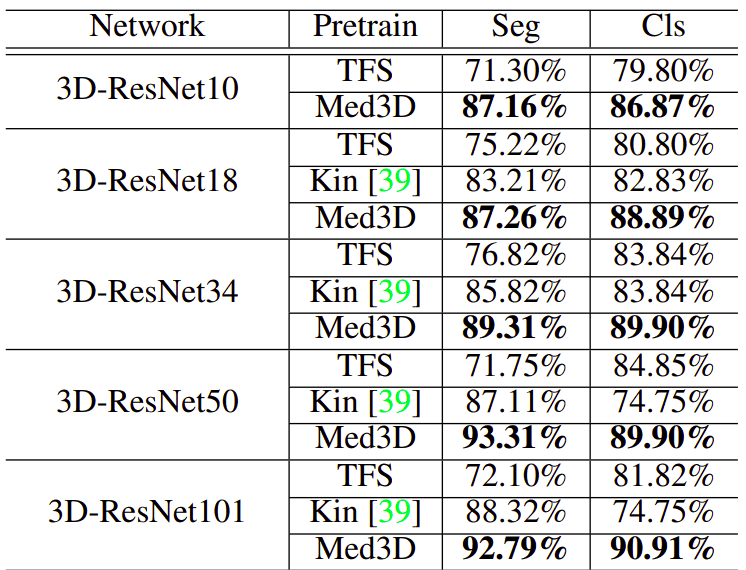 Image by Med3D: Transfer Learning for 3D Medical Image Analysis. Source
Image by Med3D: Transfer Learning for 3D Medical Image Analysis. Source
This table exposes the need for large-scale medical imaging datasets. ResNet’s show a huge gain both in segmentation (left column) as well as in classification (right column). Notice that lung segmentation exhibits a bigger gain due to the task relevance. In both cases, only the encoder was pretrained.
Teacher-Student Transfer Learning for Histology Image Classification
This is a more recent transfer learning scheme. It is also considered as semi-supervised transfer learning. First, let’s analyze how the teacher-student methods work. As you can imagine there are two networks named teacher and student. Transfer learning in this case refers to moving knowledge from the teacher model to the student. For the record, this method holds one of the best performing scores on image classification in ImageNet by Xie et al. 2020 [5]. An important concept is pseudo-labeling, where a trained model predicts labels on unlabeled data. The generated labels (pseudo-labels) are then used for further training.
The teacher network is trained on a small labeled dataset. Then, it is used to produce pseudo-labels in order to predict the labels for a large unlabeled dataset.
The student network is trained on both labeled and pseudo-labeled data. It iteratively tries to improve pseudo labels. It is a common practice to add noise to the student for better performance while training. Noise can be any data augmentation such as rotation, translation, cropping. Finally, we use the trained student to pseudo-label all the unlabeled data again.
In the teacher-student learning framework, the performance of the model depends on the similarity between the source and target domain. When the domains are more similar, higher performance can be achieved. The best performance can be achieved when the knowledge is transferred from a teacher that is pre-trained on a domain that is close to the target domain.
Such an approach has been tested on small-sized medical images by Shaw et al [7]. Specifically, they applied this method on digital histology tissue images. The tissue is stained to highlight features of diagnostic value.
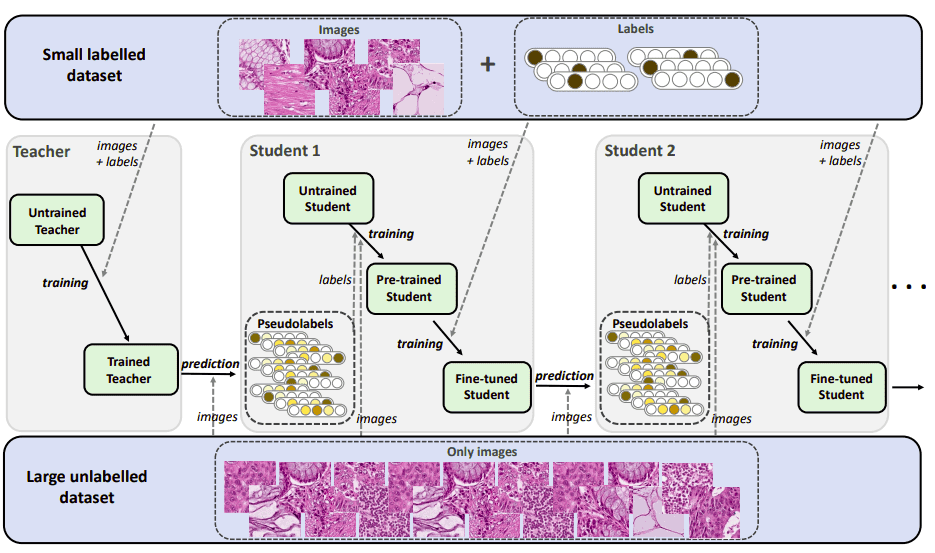 Iterative teacher-student example for semi-supervised
transfer learning. The image is taken from Shaw et al. [7]. Source
Iterative teacher-student example for semi-supervised
transfer learning. The image is taken from Shaw et al. [7]. Source
This type of iterative optimization is a relatively new way of dealing with limited labels. At the end of the training the student usually outperforms the teacher. As a consequence, it becomes the next teacher that will create better pseudo-labels. This method is usually applied with heavy data augmentation in the training of the student, called noisy student.
Conclusion
We have briefly inspected a wide range of works around transfer learning in medical images. Still, it remains an unsolved topic since the diversity between domains (medical imaging modalities) is huge. That makes it challenging to transfer knowledge as we saw. Another interesting direction is self-supervised learning. We have not covered this category on medical images yet. I hope by now that you get the idea that simply loading pretrained models is not going to work in medical images. Until the ImageNet-like dataset of the medical world is created, stay tuned. And if you liked this article, share it with your community :)
Want more hands-on experience in AI in medical imaging? Apply what you learned in the AI for Medicine course.
References
- [1] Raghu, M., Zhang, C., Kleinberg, J., & Bengio, S. (2019). Transfusion: Understanding transfer learning for medical imaging. In Advances in neural information processing systems (pp. 3347-3357).
- [2] Chen, S., Ma, K., & Zheng, Y. (2019). Med3d: Transfer learning for 3d medical image analysis. arXiv preprint arXiv:1904.00625.
- [3] Taleb, A., Loetzsch, W., Danz, N., Severin, J., Gaertner, T., Bergner, B., & Lippert, C. (2020). 3D Self-Supervised Methods for Medical Imaging. arXiv preprint arXiv:2006.03829.
- [4] Wacker, J., Ladeira, M., & Nascimento, J. E. V. (2019). Transfer Learning for Brain Tumor Segmentation. arXiv preprint arXiv:1912.12452.
- [5] Xie, Q., Luong, M. T., Hovy, E., & Le, Q. V. (2020). Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 10687-10698).
- [6][wikipedia. lung nodule](https://en.wikipedia.org/wiki/Lung_nodule)
- [7] Shaw, S., Pajak, M., Lisowska, A., Tsaftaris, S. A., & O'Neil, A. Q. (2020). Teacher-Student chain for efficient semi-supervised histology image classification. arXiv preprint arXiv:2003.08797.
* Disclosure: Please note that some of the links above might be affiliate links, and at no additional cost to you, we will earn a commission if you decide to make a purchase after clicking through.